Sensitivity and specificity answer the wrong questions
Why how we measure a diagnostic test and how we interpret them are equally important
A few weeks ago, I wrote an article on diagnostic stewardship and Bayesian reasoning when making a diagnosis. In order to keep the article brief, I stayed away from how we measure the accuracy of diagnostic tests but recently, my friend, ex-colleague and IntelliVet Founder, Katie Fox, asked me for my thoughts on some new diagnostics which led to an interesting discussion on this topic.

Fluoretiq have created novel diagnostic methods for antimicrobial susceptibility testing and identification of bacterial species in urine, the latter of which is aiming for the veterinary market. The Veri-5 Vet machine uses their Nanoplex technology which "enables point-of-care ID and enumeration to rapidly diagnose bacterial infections". Katie gave permission to mention her and share my response with you here.
My response
"The fluoretiq thing is interesting... Certainly looks promising and looks like a good team behind it.
The antibiotic susceptibility testing aspect sounds great but it's not really clear how it compares to reference/standard AST methods. Maybe it's just because they don't want to publish until the company is fully functioning but virtually all of the publications on the website are only posters or conference abstracts so the info is limited. Will be interesting to see what comes out of it and how it's validated for sure.
The vet UTI thing I'm possibly a bit more cynical about. Again all the publications [on their website] are posters using cultures or synthetic urine samples although it mentions a trial on human urine on the page but it would be good to know how it holds up with actual cat/dog urine. If it does what it says it does, then I think it really depends on cost as to how much benefit it is as I'm not sure it's solving the problem it intends to solve. Like from a stewardship point of view, having a bacterial ID would be great as K. pneumoniae and Enterobacter spp. have some intrinsic resistances that could influence empirical antibiotic choice - however, I think this gain is only marginal.
Most practices have a microscope (or another machine) to identify an active sediment and bacteria in urine, so these really need to be the reference test to compare to in my opinion. Like 50-60% of UTIs are E. coli, so empirical amoxicillin will treat most cases unless it's got acquired resistance (which this machine doesn't tell you anyway). Then the other gram negative bacteria they mention have an incidence of like up to 5% each and their sensitivity and specificity are 90% ish so the actual positive predictive value won't be great... I actually did a little calculation with 1000 UTIs to see 😂 Ignore all the half patients, I was back calculating from the sens/specificity.
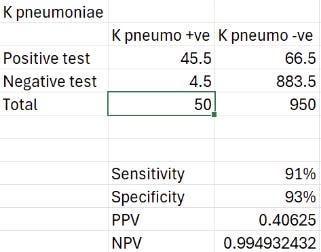
K. pneumo with 5% incidence:
Not sure I'd say 40% chance of a correct ID is worth escalating antibiotics but then of course I wouldn't! The negative predictive value is good of course, but that's probably more relevant if you want to exclude those that are susceptible to your empirical choice (i.e. wild-type E. coli). Really the specificity needs to be approaching 98-100% for me to trust the result with organisms that have a low incidence.
So all in all, I'm unconvinced. If it costs the same as a other bacterial identification methods then maybe worth it for the extra info? Still not sure I'd trust it enough to change my empirical treatment but maybe it would prompt sending off for culture and AST if something unusual comes up rather than waiting for treatment to fail if not going to culture immediately."

Sounds a bit like we’re rolling the dice on the diagnosis doesn’t it? Let’s dig a bit deeper and discuss what we are actually using a test for and how to measure its effectiveness.
Asking the right questions
To be clear, novel infection diagnostics are much needed right now and the technology behind this looks fascinating, but the clinical utility needs to be put into context. This machine from Fluoretiq answers two questions for us (partially):
Are there bacteria in this urine?
What type of bacteria are they?
Putting this into clinical context, it's really unclear how much these would change clinical practice. Being able to identify bacteria in urine is certainly useful, but we don't yet know how it compares to current in-house methods like microscopy, although this is inevitably clinician dependent. Comparing it to culture is useful but it's important to remember how this serves a fundamentally different purpose in antimicrobial stewardship by allowing antimicrobial susceptibility testing.
Even if a clinician isn't able to identify bacteriuria, this machine still doesn't fully answer whether there are bacteria in urine. It actually only identifies 4 species of Gram-negative bacteria (E. coli, K. pneumoniae, P. mirabilis and Ent. cloacae complex). Problem is, Gram-positive cocci are reported in roughly 20-30% of urine cultures and this machine has no ability to detect them (or the other remaining 10% or so of other species). So we may actually miss up to 40% of cases from the get go.
Finally, we come to the important question of whether the result would even affect your treatment. Even if this machine gave us 100% perfect answers every time, it only gives us information on bacterial species. Therefore, we can only base our antimicrobial choice on the expected (intrinsic) resistance phenotypes of the wild-type of these species. Out of our 4 identifiable species, the European Committee on Antimicrobial Susceptibility Testing (EUCAST) only categorises K. pneumoniae and Enterobacter cloacae complex as intrinsically resistant to Ampicillin/Amoxicillin which would be my first empirical drug of choice.
And this is how we end up at the prevalence problem.
The right tool for the job
Sensitivity and specificity are fundamental measurements of test accuracy. Problem is, they don't actually answer a clinically relevant question. By definition, these measures tell you how likely a test is positive in patients confirmed to have the disease of interest. Unfortunately, we don't know if a patient has the disease when we test them (that's why we do it in the first place!).
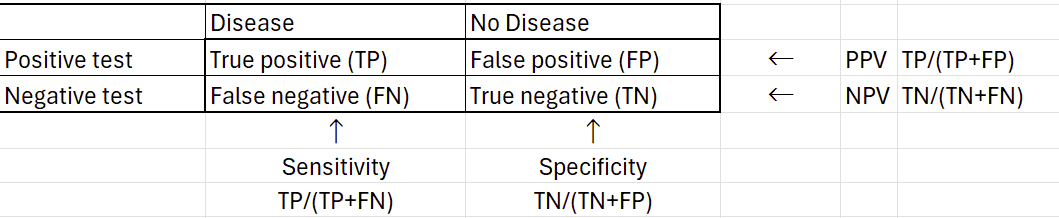
This is where positive and negative predictive values (PPV and NPV) come into play. These tell us how likely the patient has the disease given a positive/negative test. All of these measures can be calculated from our standard 2x2 contingency table:
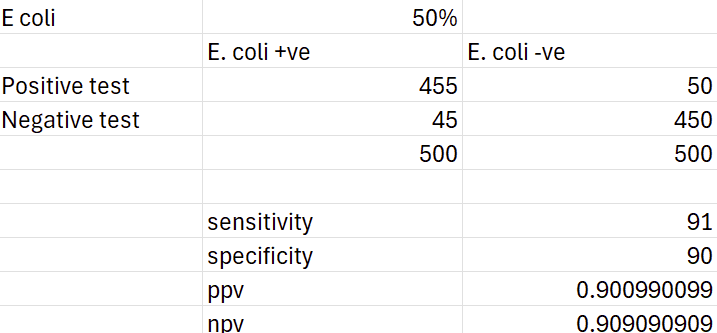
One key difference between these two measures is that predictive values massively depend on the prevalence of the disease. Interestingly, sensitivity and specificity have also been shown to vary with prevalence in epidemiologic studies even though mathematically they shouldn't. Regardless, here's the result using E. coli and a culture rate of 50%:
Clearly this is a much better result, even with a slightly lower sensitivity and specificity. Unfortunately, it is still clinically useless as we wouldn't prescribe any differently to our standard empirical choices.
Bringing it all together
Studies report culture rates of Klebsiella spp. generally around 2-3% with occasional reports around 10% or higher and Enterobacteria spp. of 1-2% although possibly up to 7% for pyelonephritis. However, these are typically reported at the genus level rather than species level, so the prevalence for K. pneumoniae and E. cloacae complex may actually be even lower.
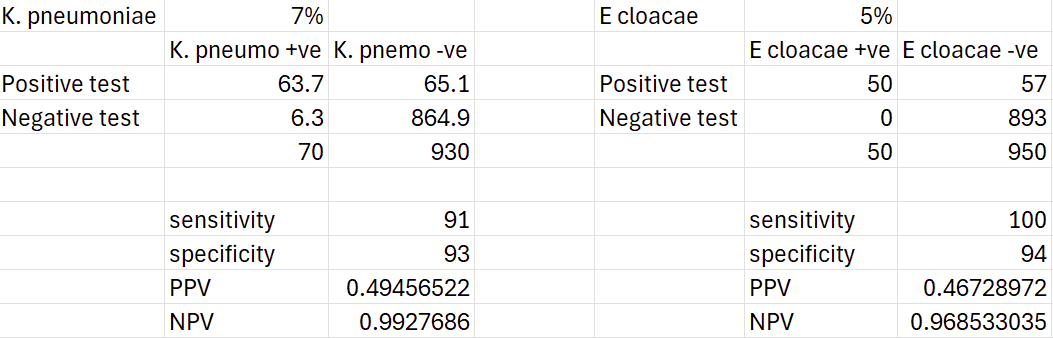
For now though, lets give these a really generous prevalence of 7% and 5% each. Out of 100 patients that come into the practice with a culture positive urinary tract infection (and assuming they are infected with a single bacterial species), 12 animals are potentially detectable and could influence our prescribing. With some updated contingency tables, we can have a crack at how many we might actually get right:
Based on these, we might correctly identify 6 K. pneumoniae cases and 5 Enterobacter cloacae cases, which is nearly all of them. On the other hand, the PPVs of 49% and 46% mean the cases that test positive are no better than a coin flip as to whether they are true positives or false positive. You could treat them all, but you’ll be giving higher-priority antimicrobials to half of the positive testing cases unnecessarily… or you could symptomatically treat and wait for a culture result in a clinically stable animal.
What's worse is that this is an ideal-world scenario. Of course we don't know who has a UTI when they walk in the door, the prevalence of these organisms is likely much lower and some infections are mixed-species, not to mention the diagnostic stewardship needed so as not to treat subclinical bacteriuria.
Conclusion
I'm interested to see where this technology goes, but right now, I'm not sure where it's place is in the veterinary practice or laboratory. I'd be curious to hear your thoughts on it so please do comment if you have a different opinion.
Otherwise, it has at least been a great example of how we measure the accuracy of a test and why sometimes we should think twice before relying on sensitivity and specificity alone. If you want to read more on this, I highly recommend Tom Boyle's How to Request a Test (affiliate link) or this article in JSAP by Brennan McKenzie.
Bibliography
Teh, H., A review of the current concepts in canine urinary tract infections. Aust Vet J. 2022; 100: 56–62. https://doi.org/10.1111/avj.13127
Wong, C., Epstein, S.E. and Westropp, J.L. Antimicrobial Susceptibility Patterns in Urinary Tract Infections in Dogs (2010–2013). J Vet Intern Med. 2015; 29: 1045-1052. https://doi.org/10.1111/jvim.13571
Garcês, A., Lopes, R., Silva, A., Sampaio, F., Duque, D., & Brilhante-Simões, P. Bacterial Isolates from Urinary Tract Infection in Dogs and Cats in Portugal, and Their Antibiotic Susceptibility Pattern: A Retrospective Study of 5 Years (2017-2021). Antibiotics (Basel, Switzerland). (2022); 11: 1520. https://doi.org/10.3390/antibiotics11111520
Windahl, U., Holst, B.S., Nyman, A. et al. Characterisation of bacterial growth and antimicrobial susceptibility patterns in canine urinary tract infections. BMC Vet Res. 2014; 10: 217. https://doi.org/10.1186/s12917-014-0217-4
McKenzie, B.A. Rational use of diagnostic and screening tests. J Small Anim Pract. 2021; 62: 1016-1021. https://doi.org/10.1111/jsap.13393